|
| |
|
| |
|
|







|
|
TCHS 4O 2000 [4o's nonsense] alvinny [2] - csq - edchong jenming - joseph - law meepok - mingqi - pea pengkian [2] - qwergopot - woof xinghao - zhengyu HCJC 01S60 [understated sixzero] andy - edwin - jack jiaqi - peter - rex serena SAF 21SA khenghui - jiaming - jinrui [2] ritchie - vicknesh - zhenhao Others Lwei [2] - shaowei - website links - Alien Loves Predator BloggerSG Cute Overload! Cyanide and Happiness Daily Bunny Hamleto Hattrick Magic: The Gathering The Onion The Order of the Stick Perry Bible Fellowship PvP Online Soccernet Sluggy Freelance The Students' Sketchpad Talk Rock Talking Cock.com Tom the Dancing Bug Wikipedia Wulffmorgenthaler |
|
bert's blog v1.21 Powered by glolg Programmed with Perl 5.6.1 on Apache/1.3.27 (Red Hat Linux) best viewed at 1024 x 768 resolution on Internet Explorer 6.0+ or Mozilla Firefox 1.5+ entry views: 1820 today's page views: 598 (20 mobile) all-time page views: 3386639 most viewed entry: 18739 views most commented entry: 14 comments number of entries: 1226 page created Fri Jun 20, 2025 14:16:12 |
|
- tagcloud - academics [70] art [8] changelog [49] current events [36] cute stuff [12] gaming [11] music [8] outings [16] philosophy [10] poetry [4] programming [15] rants [5] reviews [8] sport [37] travel [19] work [3] miscellaneous [75] |
|
- category tags - academics art changelog current events cute stuff gaming miscellaneous music outings philosophy poetry programming rants reviews sport travel work tags in total: 386 |

| ||
|
- academics - No EPL this week, so it's just my idea of leisure and me. HEp-2 Update Screening the just-released Technical Program for ICPR turned up at least three papers (and presumably contestants) on the HEp-2 classification problem described here previously. A team from Hong Kong used Multiclass Boosting SVM, though no hard figures about the results are available in their abstract, while representatives from SUNY Buffalo claimed 91.13% on HOG, texture and ROI features, also using SVM, though it was not immediately clear if this accuracy was obtained using leave-one-out. More details are accessible for the entry from Lund, Sweden, as they have very kindly placed their paper online. They report a leave-one-out accuracy of 97.4%, 1% higher than what I managed (apparently, the previous best on this dataset was 79.3%, with Adaboost), using the Random Trees classifier on close to a thousand features; unsurprisingly, the distribution of their confusion matrix is quite similar to mine, being born of the same data and uncertainties:
In both cases, the confusion between the similar-looking homogeneous and fine speckled classes is the strongest, and they have additionally improved upon some categories, most notably on the fine speckled cells. 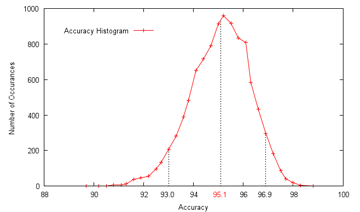 To gain some estimate of the reliability of my results, I ran my SVM-based procedure 10000 times on random even (not leave-one-out) splits of the given data, which gave an average of 95.1% accuracy, a minimum of 89.7% and a maximum of 98.9%, with a standard deviation of 1.18. 90% of the range was between 93.0% and 96.9%, with the distribution approximately symmetrical; since the actual competition held-out test set is also the same size as the given data, I would hazard that the actual spread would be comparable (±2% in a 90% confidence interval), perhaps a bit less. In any case, I believe that getting some 96% leave-one-out accuracy with just ten features might still offer some insights as to how compact an effective descriptor for the cell classes can be, again assuming no implementation errors. Go Mitt! (Yeah Right) With the deadline for the Mitosis Detection contest approaching, it was time to finally get onto it in earnest. After squinting at the dataset for ages, I had to conclude that it was far harder than the HEp-2 one, at least in terms of predicted raw classification accuracy achievable; while there are only two classes - mitotic or non-mitotic, the difference is that while I feel fairly confident of visually identifying the type of HEp-2 cells, I can't tell between mitotic and non-mitotic cells myself - so what hope of training a computer to do it? 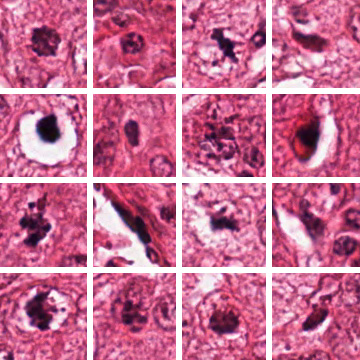 Which of these are mitotic? Since I had signed up for it, though, I felt obliged to give it a go, and therefore hardened my forehead and began to probe for clues. The first question was whether it was even possible to segment out the mitotic nuclei candidates for consideration, and happily enough a preliminary investigation revealed that they were generally far darker than the average pixel in the slide image. What this means is that, taking only the darkest 1% of pixels, they would include 47% of the pixels marked as within a mitosis, and moreover cover half of all individual mitoses by at least 50%; these figures rise as the threshold is raised, and 91% of all mitoses would be at least half covered by the darkest 4% of pixels, and 99% by the darkest 10%. In addition, this simple method of segmentation produces candidate regions - which I shall call blobs from here on - that are close to the size and appearance of the true mitotic extent, since nuclei are generally separated from other nuclei by lighter-coloured cell tissue. The tradeoff then is that while a higher threshold includes more true mitoses, they also include more false candidates - at a 4% threshold, some 323 candidate blobs are produced on average per image, with only about 6 of them true mitoses; at 10% (which was chosen), the number of candidate blobs rises to 963: 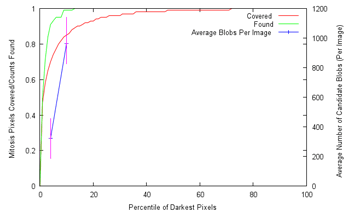 Are there any further observations that can be used to winnow more of these false candidates? Well, it can be seen that true mitoses range from 209 pixels to 2782 pixels in size, and therefore too-small blobs of about 100 pixels or below can be ignored. While about 10% of the mitoses come with multiple components, the largest of these components still has a size of over 209, so the rule still holds. Despite all this, plenty of candidates yet remain, and we shall have to, as with the HEp-2 cells, attempt to extract features from them that allow the computer to discriminate between them (as covered). From experience, the pipeline of action in such tasks is dully standard:
Size, as has been seen, is a pretty good feature, if far from enough on its own. How ideal a feature is can be estimated visually from the distributions, or slightly more rigorously using some measure of the difference in normalised distribution between the mitosis and non-mitosis classes. The results can be, and were, unexpected, with the skewedness of normalised intensity apparently being the most discriminative feature among those examined, with custom additions included after much thought, such as the average distance to neighbouring candidate blobs and the number of gaps within, being almost as useless as the position: 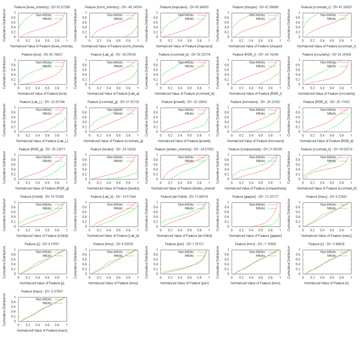 (Click to enlarge) Not that skewedness is even all that good a feature to begin with, but that's what we have got to work with. Next was throwing all of them in and having LIBSVM sort it out. Frankly, I would have been shocked had it managed to come out with anything close to the supplied opinion of human experts (which, it has to be said, do not always concur) There does not appear to have been much research down this exact line (which was part of the reason why the contest was organized), though I did dig up a fairly recent paper that had the machine agree with human experts 63.6% of the time on mitotic cells, and 98.6% of the time on non-mitotic cells, using a process that appears fairly similar to what I am attempting, with SVM as the major classification technique. These results broadly concur with my grid search findings, using ten-fold cross-validation: 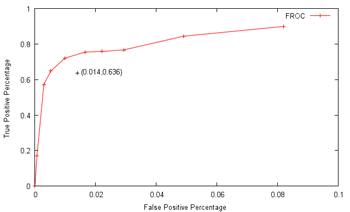 In particular, at a non-mitotic sensitivity of 98.4%, a corresponding 75.4% mitotic sensitivity is achieved, similar to the above claims (again, keep in mind that the datasets used are different). However, the problem is that there are on average over 900 candidates per image, so a 98.4% non-mitotic sensitivity, while very high on the surface, means that there would still be some 16 false positives per image! Though most of the true mitotic cells would be detected, since there are generally only like five of them per image, the output would still not be very useful for diagnostic purposes, as slides of healthy tissue would probably then return a similar number of positives as cancerous tissue. Well, I'm not gonna sweat it too much, seeing as this is just for fun, and so the final question is: what tradeoff between true and false positives should I shoot for? Now, the evaluation metrics are based on pure recall (sensitivity), precision and the F-measure, which is a combination of sensitivity and precision. Sensitivity is probably the easiest one to maximize, if other measures are disregarded - simply report all the initially-segmented blobs as true positives; recall that like 99% sensitivity is possible this way at a 10% threshold. However, the readings would then be basically completely meaningless, so I'll pass up on this slightly-cheap plan (though I wouldn't judge contestants who do go down this route for the medal) Precision is a more practical measure, which basically asks how sure one is that a candidate predicted to be mitotic by the computer actually is mitotic. Assuming that the system picks out 20 candidates, 5 of which are actually correct, the precision is then 5/20=0.25. From the graph above, one possible combination is 16.98%/99.93%, which means that the system only gets like one in six true mitoses, but almost nothing wrong, which unfortunately is again not very useful. As sensitivity and precision are both flawed, in that one can focus exclusively and score very highly on either of them while producing rubbish, it is natural to try and combine them in some way to encourage results that are fairly well-balanced. One popular metric is the F-measure, which is 2*(precision*sensitivity)/(precision+sensitivity), which seems to me like a reasonable target. With this in mind, I translated the abovementioned data points to their F-measure equivalents: 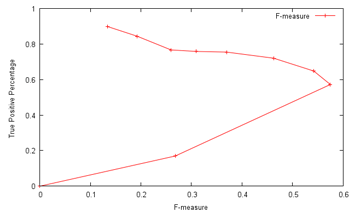 ...and the combination of 57.21%/99.70% maximizes the projected F-measure at 0.575 and indeed, detecting nearly 60% of the true mitoses while having only about three false detections per image seems kind of promising (Cohen's kappa=0.572, which seems fair). Not so bad lah. Of course, whether this performance translates over to the held-out data remains to be seen. An average of 4.2 candidates were returned from the trained model per test image, which does seem plausible enough. Well, that was disconcertingly like my usual research. Next: Downtime
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Copyright © 2006-2025 GLYS. All Rights Reserved. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||