|
| |
|
| |
|
|







|
|
TCHS 4O 2000 [4o's nonsense] alvinny [2] - csq - edchong jenming - joseph - law meepok - mingqi - pea pengkian [2] - qwergopot - woof xinghao - zhengyu HCJC 01S60 [understated sixzero] andy - edwin - jack jiaqi - peter - rex serena SAF 21SA khenghui - jiaming - jinrui [2] ritchie - vicknesh - zhenhao Others Lwei [2] - shaowei - website links - Alien Loves Predator BloggerSG Cute Overload! Cyanide and Happiness Daily Bunny Hamleto Hattrick Magic: The Gathering The Onion The Order of the Stick Perry Bible Fellowship PvP Online Soccernet Sluggy Freelance The Students' Sketchpad Talk Rock Talking Cock.com Tom the Dancing Bug Wikipedia Wulffmorgenthaler |
|
bert's blog v1.21 Powered by glolg Programmed with Perl 5.6.1 on Apache/1.3.27 (Red Hat Linux) best viewed at 1024 x 768 resolution on Internet Explorer 6.0+ or Mozilla Firefox 1.5+ entry views: 2928 today's page views: 1100 (26 mobile) all-time page views: 3733144 most viewed entry: 18739 views most commented entry: 14 comments number of entries: 1256 page created Fri Mar 6, 2026 18:18:03 |
|
- tagcloud - academics [70] art [8] changelog [49] current events [36] cute stuff [12] gaming [11] music [8] outings [16] philosophy [10] poetry [4] programming [15] rants [5] reviews [8] sport [37] travel [19] work [3] miscellaneous [75] |
|
- category tags - academics art changelog current events cute stuff gaming miscellaneous music outings philosophy poetry programming rants reviews sport travel work tags in total: 386 |

| ||
|
- academics - And so it happens that all three modules this semester happen to have a large project component. Of the three, the one on databases was the most conventional, and which for my part mostly reduced to implementing an external sort-merge in Java. Integrating code was a bit of a sweat, but we got through that. DNSRA Challenge For algorithms, we had a new project this year - a DNSRA Challenge, which goes well with the biotech direction of the local planned economy.  Making the wall of text that follows slightly more agreeable (Sources: L' esule cinefilo & Imp Awards) A teeny bit of background: Recall the big news some years ago on the (more-or-less) completion of the Human Genome Project, which aimed to produce a complete representation of human DNA. IANAB, but from introductory biology modules, DNA can be thought of as a long strand (possibly circular) of four bases - Adenine (A), Cytosine (C), Guanine (G) and Thymine (T), which is nearly completely identical for members of the same species, and often largely similar even between species. There has been much excitement about using this genetic data to understand how the human body works, especially in relation to hereditary diseases, but perhaps even abilities (see Designer Baby). For us CS people, however, this is not exactly within our scope, and the project focuses on how to obtain the genetic DNA sequence in the first place. A popular misconception might be that this is pretty straightforward - the DNA sequence is some long double-helix thingy in a cell, right? So, isn't it just a case of pulling out this thingy from a human cell, straightening it, and feeding it into some billion-dollar machine and sitting by idly, optionally humming, while waiting for the results? Well, the basic idea is there, but the problem is that we can read only a relatively tiny number of bases each time with any measure of reliability. Traditionally, this number was around a thousand bases, not a lot considering that the human genome has billions of bases. Then, a few years ago, a new method of sequencing DNA appeared on the market, which produces DNA fragments mere dozens of bases long. Wait - is this even an improvement over thousand-base fragments? It turns out that the answer is yes, if far, far more of these short fragments could be produced far more quickly and at a far lower price. Of course, as could be expected, putting these tiny fragments together becomes even more difficult than before. If assembling thousand-base fragments is akin to putting together a jigsaw puzzle of an overexposed sunset, assembling 30-base fragments might then be putting together that same jigsaw - but after dousing it in orange paint and throwing the pieces through a shredder. Strictly speaking, the assembly problem is in general NP-hard (in layman's terms, good luck solving it perfectly for large cases). However, in practice, a good-enough solution is often very useful - few refuse to take medication because one in a million people die from complications, and even the very simple heuristic of sorting the fragments by popularity, and putting them together if there are no conflicting fragments, works quite well sometimes. Our eventual solution was to analyse the output from several such heuristics (including one of our own), and then attempt to further merge the contiguous fragment output into even larger fragments. Some improvement was observed, but whether this applies generally would require far more varied test data. [Bonus tidbit for meticulous future students who just may come across this blog post while doing creative research: It is recommended to run large datasets on the Tembusu cluster, but there were no explicit directions on how to login that I could find. In the end, the method used was to ssh -t -l {username} tembusu2 from sunfire. Uploading files could then be done with wget, and downloading done by first transferring to sunfire with scp] License Plate Recognition Used SlideShare above to display our group's presentation for this, which was for the Computer Vision and Pattern Recognition module. If one thinks that license plate recognition is a rather mature technology, I would most probably agree, considering all the fines that can be collected from speed traps (then again, getting a human to pore over these images probably wouldn't cut too deeply into profits either). The professionals may have specialized equipment such as infrared cameras with super-high shutter speeds and known positions, though, while this project deals with rather more... varied conditions. The presentation should cover most of the main points adequately, but here's a bit extra on adaptive boosting (AdaBoost). The idea is similar to neural networks, as explained about this time last year - given some characteristics of an object, the computer decides how to classify it. As an example, suppose we wish to identify a critter as either a hamster or a mouse, based on its characteristics. Using the characteristic "furry" wouldn't help much, even if we were generous and chucked a few hairless mice into the mix. "Length of tail" is very promising though, and might be good enough by itself. The magic of AdaBoost is that it can combine numerous weak classifiers (such as "furry" above) into a single strong classifier, as long as the weak classifiers have a slightly better than 50% chance of making the correct prediction.  Left: Hypotheses all parallel to axis Right: Unrestricted hypotheses As an example, consider a dense bunch of blue dots concentrated at one end of a sparser spread of red dots. To generate a strong classifier, a weak hypothesis can first be guessed at at random (i.e. a line drawn), which states that all dots to one side should be blue, and all those to the other side, red. Obviously, the very first line cannot be a very good hypothesis, since no single line can divide the blue and red dots neatly. It does sets us up nicely for the key step: of increasing the weight of all dots that were classified incorrectly, so that the next weak classifier to be tried will try harder to classify them correctly. This process is repeated until the error (as measured against the sample data) becomes sufficiently small, and the (weighted) weak classifiers are then linearly combined into the final strong classifier. 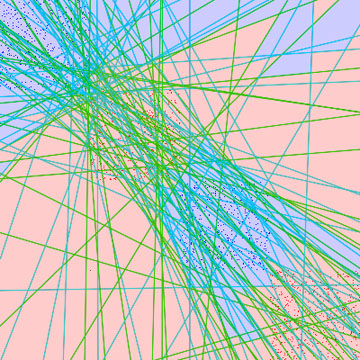 Slightly harder example That's it for the projects of this semester, and it's a relief not to have one's desktop half covered by icons any longer. Will be returning to the more usual fare soon, together with commencing revision... Next: Lie Long And Prosper
|
|||||||
 Copyright © 2006-2026 GLYS. All Rights Reserved. |
|||||||